GraphQL for the delivery of bioinformatics web APIs and application to ZincBind
This paper is a review of an emerging trend in scientific data distribution, but one that is not emerging fast enough. It is a bit different from the other papers described on this site as it is a review rather than a piece of original research, but the topic it discusses - web APIs and how they should be used to distribute scientific data - underpins a lot of modern research, especially in Bioinformatics. As always, click the link above to see the full paper.
Data Access in Science
When we think of scientific research, we might imagine teams of people working in labs, wearing lab coats, delicately transferring mysterious (and eye wateringly expensive) quantities of liquid from vial to another. Or we might think of incredibly complex (and again, staggeringly expensive) machines making strange noises as they perform some task human hands are far too clumsy to carry out.
And for the most part, that is where a lot of the science that underpins modern advances is done. But less obvious is the more intangible stuff - the data produced by all that work. Whenever an experiment is run, data is produced in the form of measurements, or analysis on those measurements, or analysis on that analysis, and so on. It's this data which lets you make inferences about the world. Without the data and its analysis, all the work done in the lab is for nothing.
But modern science is collaborative and global. If every research team produced their own data and just analysed that, we would needlessly duplicate effort on a colossal scale. We would also be limited by the ideas and expertise of the lab that produced it, rather than letting the whole world (in practice, other people in that field) examine it. Science relies on data produced by one lab being made available to anyone. And not in a 'let me know if you're interested in what we're doing and I'll email you some files when I get around to it' - systematised, reliable, automated availability.
This is a particular problem in Biology. A lot of research depends on things like sequencing entire genomes, or calculating the three dimensional structures of proteins, and that generates a lot of data - think hundreds of gigabytes at a minimum, and often much more. This is not data that you can email to your colleague in another lab as an email attachment, it is enormous. Dealing with the problem of how this data, which is central to so many other experiments, can be accessed by others, was a big headache in the 1980s and 1990s.
Part of the solution lies in having centralised organisations which act as a single data source. For example in Structural Biology there is an organisation called the Protein Data Bank. When a lab works out the 3D coordinates of a new protein, they don't invite the whole world to download that structure from them and their servers, they send it to the Protein Data Bank, and they make it available on their dedicated infrastructure.
But size is not the only constraint on making this data available. When the web started to become widely adopted in the mid-1990s, many organisations set up webpages and webforms written in HTML, where humans could navigate to a page, and manually download some data file - perhaps with a form for providing options or running a 'job' of some kind. Now anyone who wanted some data can manually go to a web page, navigate whatever links and forms are in place, and download the data files to their computer.
And this is fine if you just want that data, once. But it's a manual process. A human has to manually navigate a web page every time data is to be downloaded. Often though, you need to be able to automate this process - to download the data every few days for example, or by writing a computer program which downloads the data as part of its operation to feed into some other pipeline. Computer programs struggle to read and navigate web pages reliably, and if data was only available via web pages, this would be like doing Science with the handbrake on.
Web APIs
This is the problem that 'web APIs' solve. They work in a similar way to the normal web that humans interact with, but instead of a human typing an address into a browser and getting a HTML page back, a bot or program will request some resource at some address, and get data back in a strict format like JSON or XML, which they know how to interpret. HTML is the language that describes how web pages should look and there's a near infinite ways of laying out a page and interface. JSON and XML however are strict data descriptive languages - they can be read in an automated way by a computer program.
A common pattern that was arrived at early on was the 'REST API'. This works very similarly to how browsers work. Rather than each page having its own unique address, each 'resource' has its own unique address. So to get the data on one gene, you might send a request to something at http://api.big-organisation.org/genes/g101 where 'g101' is that gene's unique ID. Instead of getting a HTML web page back with buttons and pictures on it, you a representation of the gene in a computer-readable form such as JSON (see diagram below). A computer program can automatically send a request for this, parse the contents easily, and do whatever analysis on it that it needs to. If all it had was the HTML, it would not be able to extract the data it needed unless a human explicitly looked at the HTML first and marked down where the key information was (which could change at any time).
By the late 2010s, this is how most web APIs in Bioinformatics (and other areas) were structured. Simple REST APIs, with a fixed list of addresses that you could query. And REST is fine, mostly - nobody ever got fired for picking REST as their API architecture. But it has some serious limitations, mostly arising from that word 'fixed'.
For example, you might only need one piece of information about (to stick with our example) a gene, such as the species it comes from, but the API address for a gene always returns the same data fields every time regardless of your needs - the species it comes from, its ID, its sequence, its expression patterns, the chromosome it's on... etc. The developers who created the API can't anticipate everyone's needs, so they just have to send all the data they have and let you the user pick out what you need once its downloaded to your computer. This is called overfetching.
A related problem is what happens when you want information about multiple related objects. Genes often have annotations for example - labels for certain subsections of them - and in this simplified model there might be an address for specific annotations. If you want the full picture, you have to send a request for the gene itself, and then requests for each of those annotations belonging to it. This is called underfetching.
You might argue that this can be avoided if you would just make the annotation data part of the gene response, so that you get all the annotation data when you request the information for a gene. This solves the underfetching problem, but in so doing making the overfetching problem worse, because now people who only want a small subset of the information for a gene have to download even more data that they don't want.
These two problems, overfetching and underfetching, are irrevocably linked, in that trying to design a REST API around lessening one of them will make the other one worse. You can either break the API into lots and lots of addresses, each with a small subset of the data, and force the user to make lots of network requests, or you can have a small number of heavier addresses, which send lots of unneeded data.
And ultimately, this arises from the fact the the structure of the API is being designed at all. A team somewhere has tried to decide ahead of time how people will use their API, and designed it to best serve that. And even if they know exactly how it will be used and manage to strike the right balance for the most common use cases, there will always be people being poorly served - literally.
This is a particularly acute problem in Bioinformatics, where the underlying datasets tend to be very complex, with lots of interconnected object types with complex relationships to each other, and a wide variety of use cases for which a single API design could never hope to properly serve.
This is the problem that GraphQL solves.
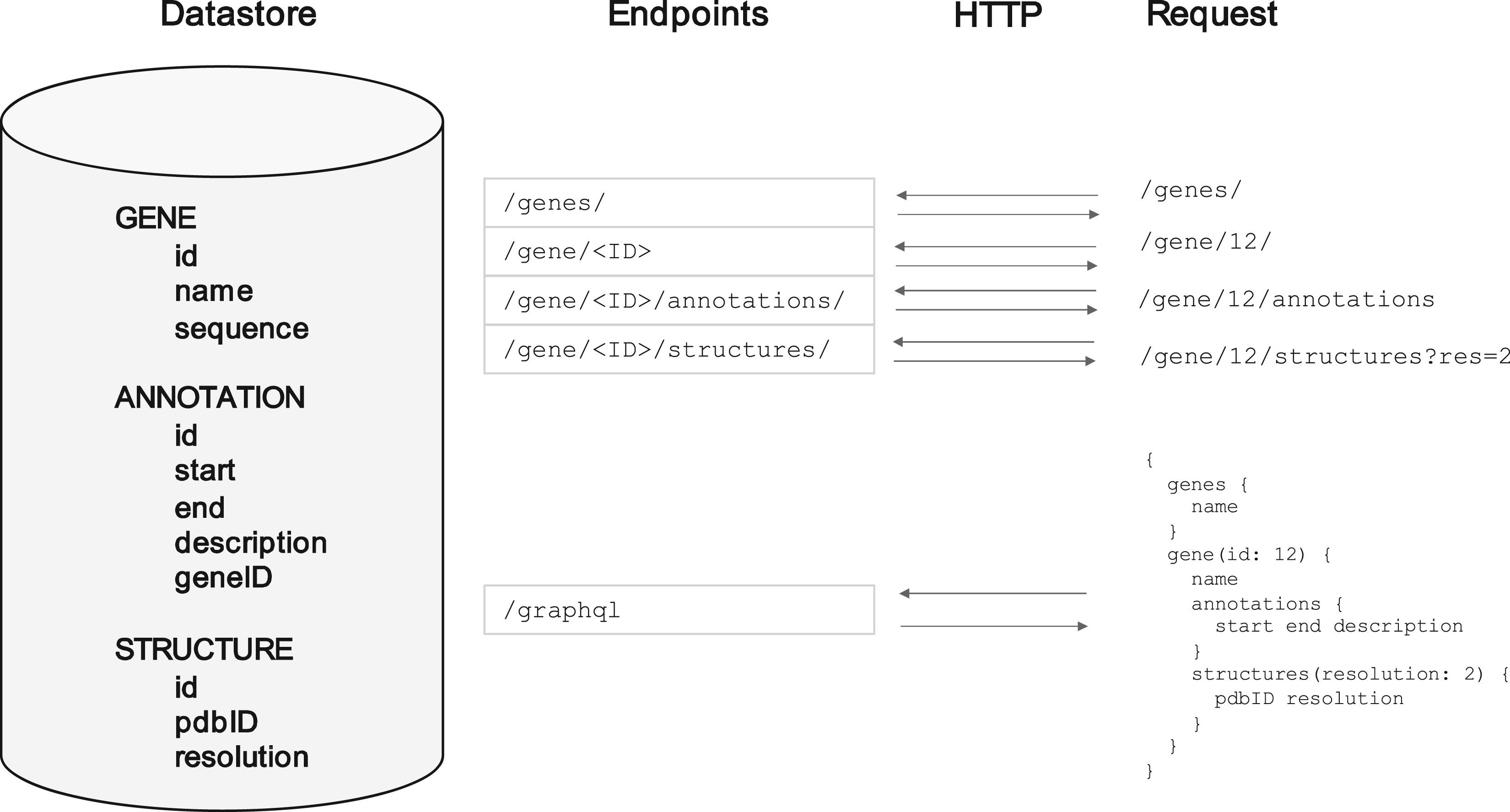
An example of a single underlying data source about genes and their annotations, and the REST API addresses that might be setup to serve it. Also shown is the equivalent GraphQL setup (see below).
GraphQL
GraphQL is a slightly different approach to providing an API. You essentially take a step back, and define a network of object types you want to make available, in a document called a schema, and then define the relationships between them. You might define a gene object type, with a name property, and an annotations property which points to the Annotation object type in a one-to-many relationship. The Annotation object type will in turn point back to the Gene object type with its gene property. This can be expanded into a whole network of object types called a graph.
You then write code which defines how each object type will return a particular property if it is asked to do so. So the gene object type will have a function called a resolver function for its annotation property which would go to some database and get all the annotations that belong to it.
Then, rather than exposing multiple pre-defined addresses, you expose a single address to which you send a query written in the Graph Query Language which asks for precisely the data you want from that graph. The API takes that query, maps it onto the previously defined network of object types and uses the resolver functions to populate that response.

The graph/network of object types in the ZincBind API.
While a little more complex to get your head around at first, and a little more work for the developers of the API, this is more than made up for in the power it gives the users. You can specify exactly what data you want, and get it in a single request to the server. You don't have to pick from a pre-defined list of addresses/response types, because nobody has tried to anticipate what you will need or required you to fit into a neat little box.
GraphQL was developed by Facebook and released in 2015, and since then has seen increasing adoption - but not in Bioinformatics much (though this does seem to be changing very recently). Bioinformatics resources tend to be slow to adopt new practices, but it in particular stands to gain a lot from increased adoption of GraphQL. It will make research and finding new discoveries just that little bit easier.
And that's partly why we wrote this review - GraphQL's benefits are not yet widely known among Bioinformaticians, and the sooner they are, the sooner we all benefit. The paper itself goes into more technical detail, as we also describe how we switched our own ZincBind API from REST to GraphQL in 2020. This is the future of Bioinformatics APIs - hopefully.