The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new guide to IMMUNOPHARMACOLOGY
I was only a minor contributor to this paper, though it is for some of the first proper academic work I ever did.
The Guide to Pharmacology is a database of drugs, and the protein targets that they act on. It’s a very useful pharmacological resource, and what's more it's a manually curated database, which means that it is built and maintained by expert pharmacologists who update the database based on the latest research and publications. They publish updates to it every few months.
The Guide's homepage
The team that operates the database is based in Edinburgh, in Prof. Jamie Davies' lab, and from September 2015 to September 2016, I joined the team to work on the construction of a new database, based on the Guide, which would use information from the Guide to offer information for Synthetic Biologists.
That new database that I ended up building is a story for another day and another paper, and it’s not why my name is on this paper - but it is why, in September 2015, I joined the team. And as an initial project, it was suggested that I could build a tool for the main database itself first, before starting work on the new system - both as a means of getting to grips with the database and the overall system, and because, well, it needed something in particular - a BLAST search tool. That’s what I built in September and October 2015, and that’s my contribution to this paper. The paper chronicles all the major advancements and improvements made to the Guide to Pharmacology site in the last two years, including the addition of this BLAST tool which I created.
Searching for Sequences
So what is a BLAST search tool, and why is it needed? What problem does it solve?
As mentioned, the database is, in essence, a list of two principle things - drugs, and drug targets. Most people who use the site don’t browse through these lists looking for the thing they want, they search the database using some attributes. For example, they might search the drugs to get those under a certain weight, or which act on a certain organ. Or, they might search the drug targets for those from a particular species or belonging to a particular class of proteins. The point is that you can search by these criteria because the database stores information about these attributes.
One attribute of drug targets that is not stored in the database is what is called protein sequence.
Essentially proteins - the molecules in living things that carry out essentially every molecular function - are chains of smaller molecules called amino acids. These amino acids might be just 5-10 atoms in size, but a protein might have 300 of these chained together in a row, making proteins rather large molecules. Rather conveniently there are only 20 distinct amino acids used by living things to make the huge variety of proteins in existence - just as the 26 letters of the alphabet can be combined to make a large variety of words. In fact, proteins are often described in just such a way. Each of the amino acids have full names like ‘glutamate’ and ‘histidine’ but they are also all given a single letter code. So you can describe the full sequence of atoms in a protein simply by writing a string of letters like MTFFPAH... This protein sequence doesn’t tell you how the atoms are arranged three dimensionally, but it tells you what amino acids the protein is made of, and the order they are in.
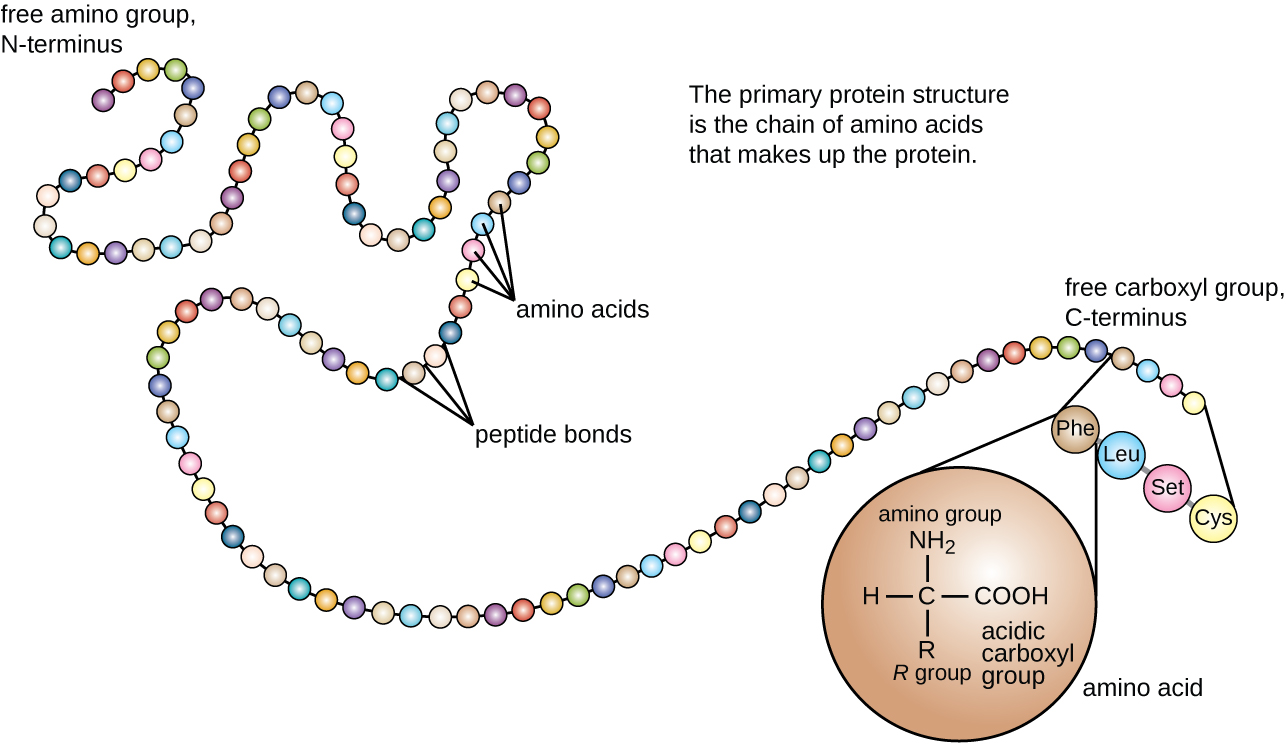
A helpful illustration of this principle that I stole from wikipedia - here three-letter codes are used to represent each amino acid, not single letters, but the principle is the same
The drug targets in the guide are all proteins, so they each have a protein sequence - though the Guide doesn't actually store this. So if you are studying a protein that you have just isolated in the lab, and you know its sequence and want to see if any drugs interact with your protein of interest, a tool which lets you search the Guide to Pharmacology by that might be quite useful, and indeed this is one thing that a BLAST search does.
However, in most circumstances, such a straightforward search isn’t actually that useful. Inputting a protein sequence, or section of one, will only return entries that are exact matches, or which contain the sequence you entered exactly. That is, you will only get a match if the protein you are studying is already in the database. But in that case you can just search by name to see if your protein is already in the database, as you will probably know the name.
What you really need, is a search tool that will return entries that are ‘a bit like’ your protein sequence - drug targets which have a very similar sequence to the one you are studying. That way, even if your protein isn't a known drug target and isn't in the database, you can see if there any known drug targets which have a similar sequence - and proteins with similar sequences tend to have similar structures and sometimes even bind to the same molecules.
(It can also give information about evolutionary relatedness, but we can put that to one side for now. The point is that such a search tool would tell you if there are drug targets that are ‘close’ to your own protein that you are working with.)
Normal database searches won’t do this. They only know how to look for exact textual matches. There are a number of ways of determining how ‘close’ two pieces of text (which is what the protein sequences are being processed as) are, but when it comes to Biological sequences like protein sequences, BLAST is king.
BLAST
It's worth stopping to ask what such a sequence search tool needs to be able to do. Suppose you input the sequence ABCDEF, and it searches the database. It needs to be able to match something like ZABCDEFY, where there is an exact match. But what about, say, ABCZDEF? Here there has been a character inserted into the string, but we can intuitvely see that it is 'close' to the search string - even if we couldn't say specifically how close.
Well, the BLAST alogorithm can. It is designed to 'align' a query string with a bunch of target strings (it stands for Basic Local Alignment Search Tool) to show which characters match up and, crucially, give you a numeric measure of how closely they match.
When BLAST is used to search the database, it can return the matching sequences in the order of closeness (which might rouhgly be expected to correspond to how closely they are related to the target sequence, with very close matches probably being structurally similar too).
What's more, it doesn't just treat the sequences as generic strings, but treats them as amino acids. That is, it knows which amino acids are 'similar' to each other, and can take this into consideration when evaluating how much two sequences match. This means that you have to have different variants of BLAST when dealing with protein sequences and DNA sequences (whose single letter codes mean entirely different building blocks) however.
Because of this, and crucially because of its speed (it takes a few shortcuts that previous algorithms had not, which made them take forever when searching large databases), BLAST is now the standard way to search lists of sequences for matches.
Building the Tool
Fortunately, I didn't actually have to implement BLAST myself or code it from scratch - the NCBI very kindly provide command line programs which will search a list of sequences for you. The challenge was (1) actually getting the protein sequences of all the drug targets in the Guide, and (2) making this command line tool usable from the website itself.
The first problem was reasonably straightforward to solve. While the Guide doesn't store sequence information, it does store database links - the IDs of each target in other database. For example, there is another database out there called Uniprot, and if a drug target in the Guide has an entry in Uniprot, its Uniprot ID will be stored in the Guide. There are many other databases which the Guide links to, but since Uniprot is a database of protein sequences, this was the crucial one. I was able to write a Python script which went through all the drug targets in the Guide, made a note of the Uniprot ID if it had one, and then used that to get the full protein sequence for the target. These were then placed into a single .fasta file (the standard file format for storing biological sequences) - essentially a separate database in a single file, which referred back to the main database.
The second problem was more fiddly. The BLAST program would live in the website's server, and generally expects to be called by a person manually entering a command into a terminal. What was needed here was a way for the Guide web app to be able to call the program, and read the results back. To cut a long story short this meant using Java (the programming language the Guide is built in) to call the BLAST program with the query it had received, get the program to save its results to a temporary file, read the contents of the file, and then delete the temporary file. A bit circuitous, but the fastest way, it turned out.
Everything else was just a matter of sticking the various components together, and integrating it with the main Guide to Pharmacology website - which at the time was a fairly intimidating prospect, given that I had been there barely a month.
But, slightly to my own surprise, the end result worked perfectly. You can try it out here - there are a few details I left out, such as the two different BLAST programs needed to also search by DNA sequence, but that's the gist of what I did here.
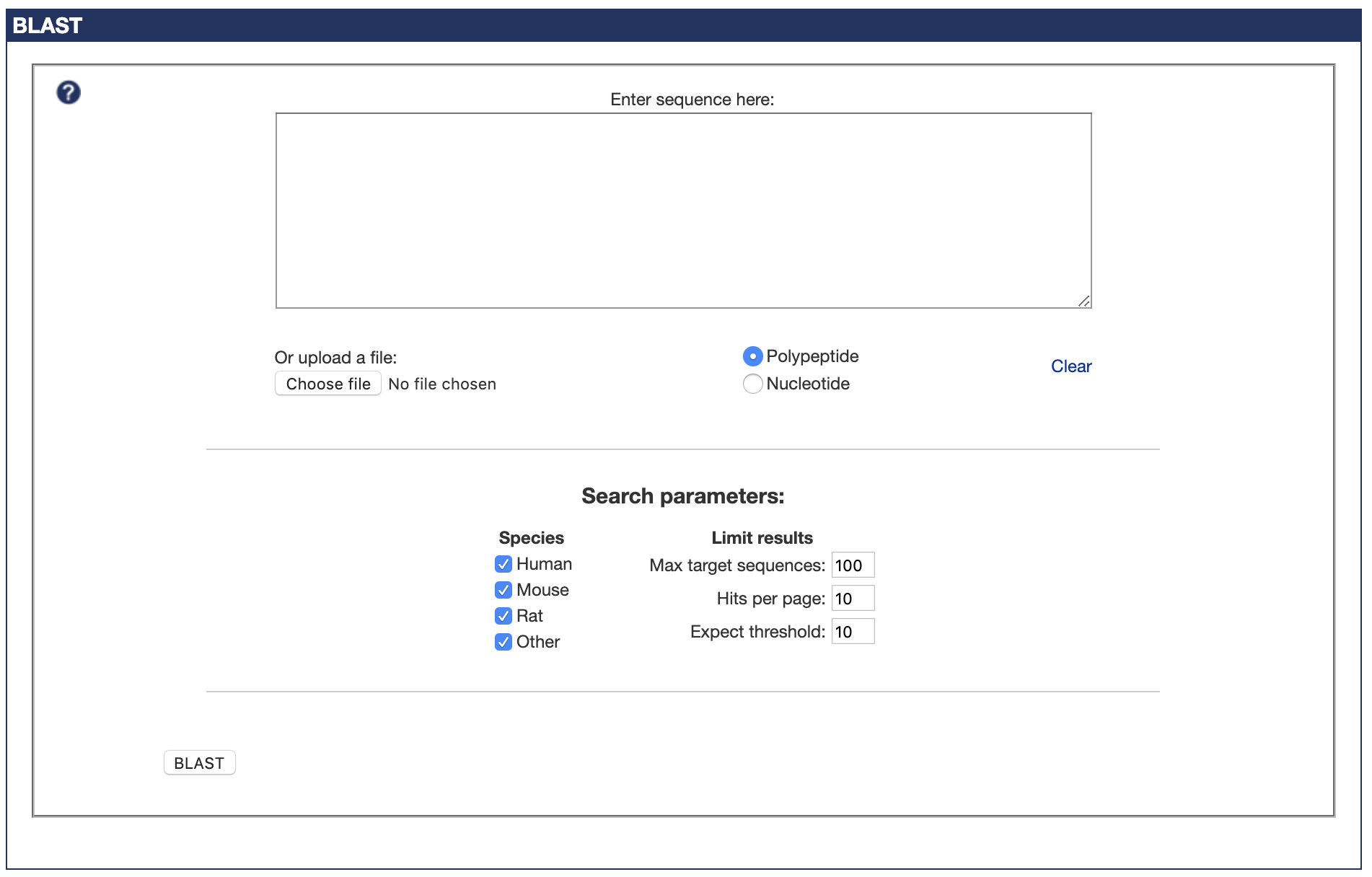
The fruit of my labours
I'm hugely grateful to the Guide to Pharmacology team for helping me get to grips with the system when I first arrived - especially Dr. Jo Sharman, who was very patient and helpful as I gradually learned how their system was set up, and worked my way towards building this system.
Anyway, that's why my name's on the paper.